Image Registration Method 3
Overview
If you are not familiar with the SimpleITK registration framework we recommend that you read the registration overview before continuing with the example.
Example Run
Running the Python code with the following inputs:
main('BrainProtonDensitySliceBorder20.png', 'BrainProtonDensitySliceShifted13x17y.png', 'displaceMeth3.hdf5')
produces the text and images below.
Output Text
Text Output (click triangle to collapse)
Estimated Scales: (29682.41582933708, 29682.168476696, 1.0000000000010232, 0.999999999998181)
0 = -0.99671 : (1.0001225188582936, -4.8155319621057874e-05, 14.24580144271548, 18.231972935092553)
1 = -0.90442 : (1.0001395698120492, -5.723494632868289e-05, 13.436891660253242, 17.644040146872914)
2 = -0.97294 : (1.0000524825397714, -3.538901200507155e-05, 12.784457666827517, 16.886194597858392)
3 = -0.99664 : (1.000078552246351, -3.493898453141627e-05, 13.248020397809663, 17.07357013992443)
4 = -0.99617 : (1.0000080585357958, -3.574957852612509e-05, 13.003964467172034, 17.01937883295938)
5 = -0.99998 : (0.9999690242080485, -8.938378761798914e-06, 12.941455002408405, 16.77731982244243)
6 = -0.99743 : (0.9999729946034386, -1.4195394490341945e-05, 12.980498953463153, 16.89606564275891)
7 = -0.99946 : (0.9999822072360259, -1.9442192886012526e-05, 13.009300985166485, 17.01770216218198)
8 = -0.99998 : (1.0000081565952237, -1.536015920575109e-05, 12.97497046153363, 16.965475115002747)
9 = -0.99990 : (1.000006961859454, -1.5839846033647187e-05, 12.996178272885958, 16.98842704816722)
10 = -0.99999 : (0.999997104263667, -1.5781032972153657e-05, 13.008157869541614, 17.01728968260679)
11 = -0.99998 : (0.9999981089260577, -1.480066274538104e-05, 13.000149494110696, 17.003873030789478)
12 = -1.00000 : (1.0000035245702283, -1.1964202117479094e-05, 12.999557108219037, 16.988259265497895)
13 = -0.99999 : (1.0000015232779387, -1.2092316764425097e-05, 12.999892054939323, 16.996064581822232)
14 = -1.00000 : (0.9999991283248564, -1.1669994922994523e-05, 13.00013101903772, 17.003873425942555)
15 = -1.00000 : (0.9999999572679382, -1.1067338273415268e-05, 12.99999566483145, 16.99996952183884)
16 = -1.00000 : (1.0000015262190347, 2.7428357741776532e-05, 13.001481019896447, 17.00123717053477)
17 = -1.00000 : (1.0000010761172178, 2.70338949452921e-05, 13.00064749562188, 17.000728337318556)
18 = -1.00000 : (1.0000004942557568, 2.6224543927636213e-05, 12.999879933384591, 17.000124581130446)
19 = -1.00000 : (0.9999997860745542, 2.334309634129523e-05, 13.000293057931712, 16.99986431610512)
20 = -1.00000 : (0.9999997971406921, 2.288036936123565e-05, 13.000070149678, 16.99996389693141)
21 = -1.00000 : (0.9999999481117745, 2.1399925381340785e-05, 12.999868757877147, 17.000101896108283)
22 = -1.00000 : (0.999999997953508, 2.0861222466294866e-05, 12.999980029544846, 17.000051701641695)
23 = -1.00000 : (1.0000000486509963, 1.633859430051924e-05, 13.000031070402398, 16.999940906626833)
-------
itk::simple::Similarity2DTransform
Similarity2DTransform (0x56305fc2d580)
RTTI typeinfo: itk::Similarity2DTransform<double>
Reference Count: 3
Modified Time: 2649
Debug: Off
Object Name:
Observers:
none
Matrix:
1 -1.63386e-05
1.63386e-05 1
Offset: [13.0022, 16.9981]
Center: [111.204, 131.591]
Translation: [13, 16.9999]
Inverse:
1 1.63386e-05
-1.63386e-05 1
Singular: 0
Angle = 1.63386e-05
Scale =1
Optimizer stop condition: RegularStepGradientDescentOptimizerv4: Step too small after 24 iterations. Current step (6.10352e-05) is less than minimum step (0.0001).
Iteration: 25
Metric value: -0.9999999568784146
Input Images
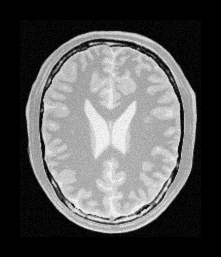
Fixed Image |
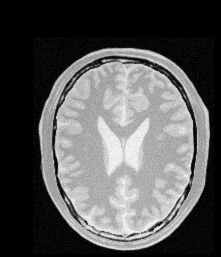
Moving Image |
Output Image
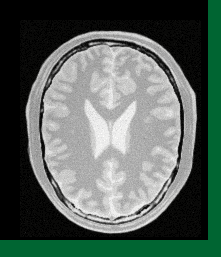
Composition Image
Code
#!/usr/bin/env python
import SimpleITK as sitk
import sys
import os
def command_iteration(method):
if method.GetOptimizerIteration() == 0:
print("Estimated Scales: ", method.GetOptimizerScales())
print(
f"{method.GetOptimizerIteration():3} "
+ f"= {method.GetMetricValue():7.5f} "
+ f": {method.GetOptimizerPosition()}"
)
def main(args):
if len(args) < 3:
print(
"Usage:",
"ImageRegistrationMethod3"
"<fixedImageFilter> <movingImageFile> <outputTransformFile>",
)
sys.exit(1)
fixed = sitk.ReadImage(args[1], sitk.sitkFloat32)
moving = sitk.ReadImage(args[2], sitk.sitkFloat32)
R = sitk.ImageRegistrationMethod()
R.SetMetricAsCorrelation()
R.SetOptimizerAsRegularStepGradientDescent(
learningRate=2.0,
minStep=1e-4,
numberOfIterations=500,
gradientMagnitudeTolerance=1e-8,
)
R.SetOptimizerScalesFromIndexShift()
tx = sitk.CenteredTransformInitializer(fixed, moving, sitk.Similarity2DTransform())
R.SetInitialTransform(tx)
R.SetInterpolator(sitk.sitkLinear)
R.AddCommand(sitk.sitkIterationEvent, lambda: command_iteration(R))
outTx = R.Execute(fixed, moving)
print("-------")
print(outTx)
print(f"Optimizer stop condition: {R.GetOptimizerStopConditionDescription()}")
print(f" Iteration: {R.GetOptimizerIteration()}")
print(f" Metric value: {R.GetMetricValue()}")
sitk.WriteTransform(outTx, args[3])
resampler = sitk.ResampleImageFilter()
resampler.SetReferenceImage(fixed)
resampler.SetInterpolator(sitk.sitkLinear)
resampler.SetDefaultPixelValue(100)
resampler.SetTransform(outTx)
out = resampler.Execute(moving)
simg1 = sitk.Cast(sitk.RescaleIntensity(fixed), sitk.sitkUInt8)
simg2 = sitk.Cast(sitk.RescaleIntensity(out), sitk.sitkUInt8)
cimg = sitk.Compose(simg1, simg2, simg1 // 2.0 + simg2 // 2.0)
return {"fixed": fixed, "moving": moving, "composition": cimg}
if __name__ == "__main__":
return_dict = main(sys.argv)
if "SITK_NOSHOW" not in os.environ:
sitk.Show(return_dict["composition"], "ImageRegistration3 Composition")
# Run with:
#
# Rscript --vanilla ImageRegistrationMethod3.R fixedImageFilter movingImageFile outputTransformFile
#
library(SimpleITK)
commandIteration <- function(method)
{
if (method$GetOptimizerIteration()==0) {
cat("Estimated Scales:", method$GetOptimizerScales())
}
msg <- paste(method$GetOptimizerIteration(), "=",
method$GetMetricValue(), ":",
method$GetOptimizerPosition(), "\n" )
cat(msg)
}
args <- commandArgs( TRUE )
if (length(args) != 3) {
stop("3 arguments expected - fixedImageFilter, movingImageFile, outputTransformFile")
}
pixelType <- 'sitkFloat32'
fixed <- ReadImage(args[[1]], 'sitkFloat32')
moving <- ReadImage(args[[2]], 'sitkFloat32')
R <- ImageRegistrationMethod()
R$SetMetricAsCorrelation()
R$SetOptimizerAsRegularStepGradientDescent(learningRate=2.0,
minStep=1e-4,
numberOfIterations=500,
relaxationFactor=0.5,
gradientMagnitudeTolerance=1e-8 )
R$SetOptimizerScalesFromIndexShift()
tx <- CenteredTransformInitializer(fixed, moving, Similarity2DTransform())
R$SetInitialTransform(tx)
R$SetInterpolator('sitkLinear')
R$AddCommand( 'sitkIterationEvent', function() commandIteration(R) )
outTx <- R$Execute(fixed, moving)
cat("-------\n")
outTx
cat("Optimizer stop condition:", R$GetOptimizerStopConditionDescription(), '\n')
cat("Iteration:", R$GetOptimizerIteration(), '\n')
cat("Metric value:", R$GetMetricValue(), '\n')
WriteTransform(outTx, args[[3]])